Primary Aim
To investigate whether taking birth control is associated with vaginal inflammation throughout the menstrual cycle.
This is an observational study to evaluate the relationship between birth control and vaginal inflammation In response to menstruation. Absolute abundance of bacteria was measured by 3 qPCR assays (for total Bacteria, L. crispatus, L. iners). Cytokine levels (in copies/ml of vaginal fluid) were also measured by Luminex. We also looked at the number and type of immune cells in the vagina using Flow Cytometry. Data was collected at four timepoints; before, at the start, the end, and after menstruation.
To investigate whether taking birth control is associated with vaginal inflammation throughout the menstrual cycle.
library(tidyverse)
library(tableone)
library(cowplot) # load for the theme
theme_set(theme_cowplot(16) + background_grid()) # set cowplot theme
sample_map <- read_csv("data/00_sample_ids_period.csv")
metadata <- read_csv("data/01_participant_metadata_period.csv")
luminex <- read_csv("data/02_luminex_period.csv")
flow_cytometry <- read_csv("data/03_flow_cytometry_period.csv")Let’s create table one
processed_for_tableone <- merged_dataset %>%
select(-limits, -sample_id) %>%
pivot_wider(names_from = cytokine, values_from = conc) %>% # move cytokine to wide
filter(time_point == "week_prior") # we just want baseline characteristics for table 1
CreateTableOne(
data = processed_for_tableone,
vars = c("age", "pcos_status", "period_product", "cd4_ratio", "IL-1a", "IL-1b", "IP-10", "MIG", "TNF-a" ),
factorVars = c("pcos_status","period_product" ),
strata = "arm",
test = FALSE
) %>%
print(
nonnormal = c("cd4_ratio", "IL-1a", "IL-1b", "IP-10", "MIG", "TNF-a"),
) Stratified by arm
birth_control no_birth_control
n 15 12
age (mean (SD)) 30.80 (2.54) 32.08 (3.20)
pcos_status = pcos (%) 6 (40.0) 8 (66.7)
period_product (%)
menstrual_cup 2 (13.3) 2 (16.7)
pad 3 (20.0) 1 ( 8.3)
tampon 10 (66.7) 9 (75.0)
cd4_ratio (median [IQR]) 0.51 [0.47, 0.52] 0.45 [0.38, 0.52]
IL-1a (median [IQR]) 25.65 [11.97, 44.53] 40.94 [11.66, 87.95]
IL-1b (median [IQR]) 0.24 [0.24, 2.04] 0.24 [0.24, 4.91]
IP-10 (median [IQR]) 11.80 [6.71, 45.20] 16.95 [6.38, 84.45]
MIG (median [IQR]) 151.24 [47.17, 301.19] 129.69 [18.63, 365.59]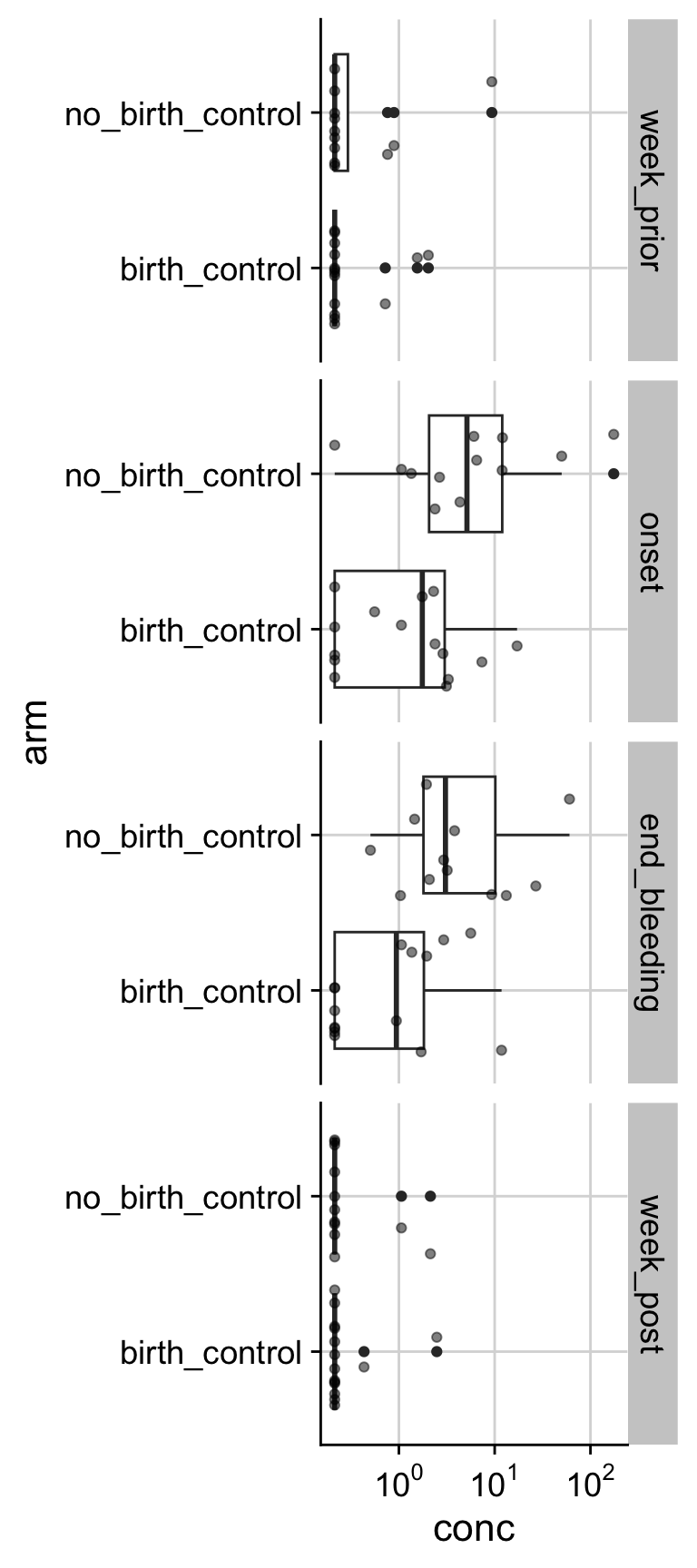
How does PCOS associate with inflammation over menstruation? First, let’s look at one of the cytokines at one of the timepoints.
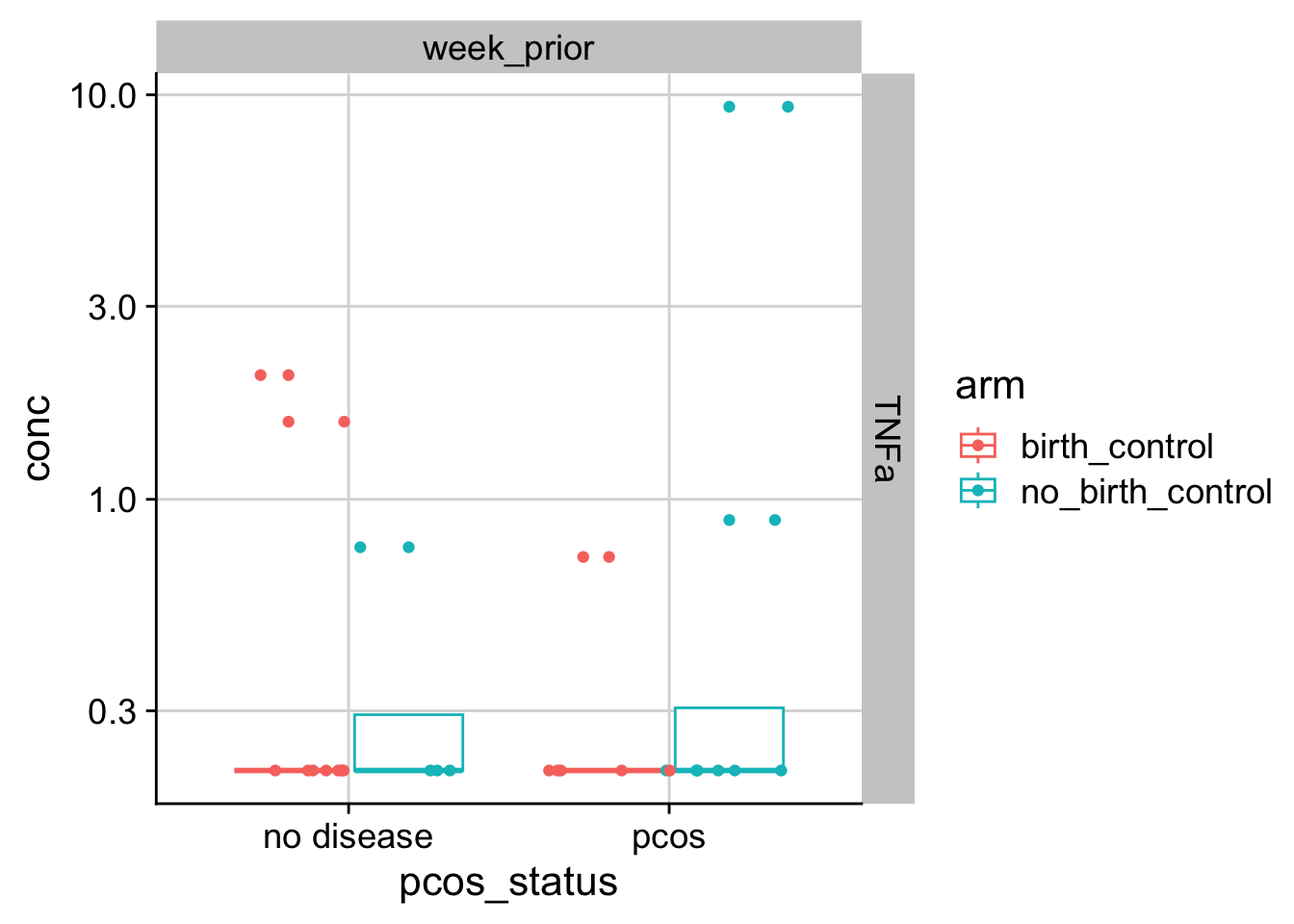
Now, let’s look at all the cytokines together with all time points.
Are there any relationships between cytokines and cd4 ratio?